Mining the equine gut metagenome: poorly-characterized taxa associated with cardiovascular fitness in endurance athletes | Communications Biology - Nature.com
Abstract
Emerging evidence indicates that the gut microbiome contributes to endurance exercise performance. Still, the extent of its functional and metabolic potential remains unknown. Using elite endurance horses as a model system for exercise responsiveness, we built an integrated horse gut gene catalog comprising ~25 million unique genes and 372 metagenome-assembled genomes. This catalog represents 4179 genera spanning 95 phyla and functional capacities primed to exploit energy from dietary, microbial, and host resources. The holo-omics approach shows that gut microbiomes enriched in Lachnospiraceae taxa are negatively associated with cardiovascular capacity. Conversely, more complex and functionally diverse microbiomes are associated with higher glucose concentrations and reduced accumulation of long-chain acylcarnitines and non-esterified fatty acids in plasma, suggesting increased ß-oxidation capacity in the mitochondria. In line with this hypothesis, more fit athletes show upregulation of mitochondrial-related genes involved in energy metabolism, biogenesis, and Ca2+ cytosolic transport, all of which are necessary to improve aerobic work power, spare glycogen usage, and enhance cardiovascular capacity. The results identify an associative link between endurance performance and gut microbiome composition and gene function, laying the basis for nutritional interventions that could benefit horse athletes.
Introduction
Endurance athletes undergo prolonged cardiovascular exercise and withstand physiological stress that disrupts the body's homeostasis. This, in turn, overwhelms organs and the system's normal function1,2. The ability to run for long distances at high speed is an uncommon feat for land mammals. Through years of selective breeding for athletic performance and tailored training practices, Arabian horses have gained built-in biological mechanisms to compete at distances of up to 160 km in a single day, an effort comparable to a human marathon or ultra-marathon runners3. Like humans4, these equine athletes display well-adapted physiological abilities, including processing large volumes of oxygen required for aerobic metabolism and markedly larger hearts than those less physically active. Unique to horses is their quadrupedal nature of locomotion, the coupling of respiration to stride frequency, and the pronounced increase in pulmonary artery pressure. These equine-specific features place different physiologic loads on the heart during endurance than those observed in humans4.
Endurance exercise performance primarily depends on cardiovascular fitness, exercise economy, and the ability to sustain both the metabolic and thermoregulatory demands of such activity4,5 without the accumulation of exponential levels of blood lactate or skeletal muscle fatigue6,7,8. Athletes with the greatest improved cardiovascular fitness and fatigue resistance often succeed in competitions4,9,10.
Undoubtedly, endurance exercise performance entails complex multifactorial processes whose mechanisms are still not fully understood. New evidence has shown that the gut microbiome and its associated metabolites can act locally in the intestine or accumulate in different body fluids11 that impact host athletic performance during endurance racing12,13. Such gut microbiome-derived metabolites include short-chain fatty acids, dimethyl sulfone, trimethylamine oxide, indoles, tryptamine, oligosaccharides, peptidoglycans, and secondary bile acids14,15, all of which affect host health16. Within the context of exercise, the gut microbiome metabolites support multiple physiological strands, e.g., energy metabolism, hydration, redox reactions, and immune responses, that can affect fatigue and stress perception2,15,17,18,19,20. For example, Veillonella atypica likely enhances athletic endurance performance via the utilization of host lactate and the production of propionate12,13. Beyond microbial-derived metabolites, changes in the microbial composition and increased diversity correlate with improved performance and cardiorespiratory fitness in marathon runners regardless of sex, age, body mass index, and diet21. Increases of the Firmicutes-Bacteroidetes ratio22, or depletion in Eubacterium spp.23 enhanced the cardiovascular capacity of athletes, as assessed by the maximum oxygen consumption.
Delineating of the relationships between the gut microbiome and endurance performance is in its infancy in humans, and it is hampered by the appropriate control of known confounding factors (such as diet, training loads, medications, occurring illnesses, environment, and genetic background). In this respect, Arabian horses emerge as a suitable in vivo model for studying the microbiome adaptations in response to endurance exercise. Because of apparent differences in anatomy—e.g., the horse cecum is large relative to the total gastrointestinal tract—and physiology, the relevance, and translation of the observations made in horses to human dietary applications may not be straightforward. However, the natural aptitude of Arabian horses for athletic performance, and the homogeneity of their genetic and environmental backgrounds, offer a more tractable system. The interdependence of exercise performance and gut microbiota in horses is underscored by several lines of evidence19,20,24,25,26,27, although the range and extent of this interplay are mainly unknown. Recent findings suggest that gut microbial metabolites in endurance horses, e.g., acetate, valerate, dimethyl sulfone, trimethylamine oxide, formate, and secondary bile acids coupled with circulating free fatty acids regulate mitochondrial function by preventing hypoglycemia25, which is the limiting factor for fatigue onset and, thus, athletic performance. Despite these findings, if and how gut microbiome functions are responsible for better adaptations to resist fatigue and succeed in athletic performances are unresolved.
To address this knowledge gap, we have built an extensive gene catalog of the gut microbiome in elite endurance horses. This expands the current representation of the equine gut microbiome with more than 25 million non-redundant genes identified and 369 new metagenome-assembled genomes (MAGs). Moreover, holo-omics data integration from the host and microbiome domains show that gut microbiome composition, functions, and mitochondria activity are critical determinants for cardiovascular fitness. Relatively poorly-described genera and their pool of genetic resources likely regulate metabolic pathways to fine-tune mitochondrial function and enhance cardiovascular capacity. On the contrary, microbial communities with reduced diversity but a higher abundance of core taxa from the Lachnospiraceae family are associated with poorer performance.
Results
Building a horse gut microbiome gene catalog and MAG repertoire
We constructed a microbial gene catalog from the feces of 11 highly trained endurance horses (Supplementary Data 1). After quality filtering and host sequence decontamination, 1124 million high-quality paired reads were available, with an average sequencing depth per sample of 93–107 million paired reads, similar to that used for the construction of the chicken28 and bovine29 gut gene catalogs (Supplementary Data 2). These data were de novo assembled (total assembly size of 21.68 Gb) to build a non-redundant gene catalog of 25,250,066 genes with an average length of 618 bp (Supplementary Note, Supplementary Data 2, Supplementary Fig. 1a–e). Individual horses harbored around half of these genes (n = 11,809,713; Fig. 1a). The core group of genes present in all individuals consisted of <7.2% of the overall microbial gene pool (n = 1,805,693; Fig. 1b). Yet, these core genes showed highly conserved abundance rank structure across individuals, representing 29.7 to 38.4% of the overall microbiome abundance. Only 22.5% (n = 922,362) of the recently published horse gut microbiome gene catalog30 overlapped with our gene catalog, indicating that the functional potential of gut microbiomes in horses is vast and currently under-sampled.
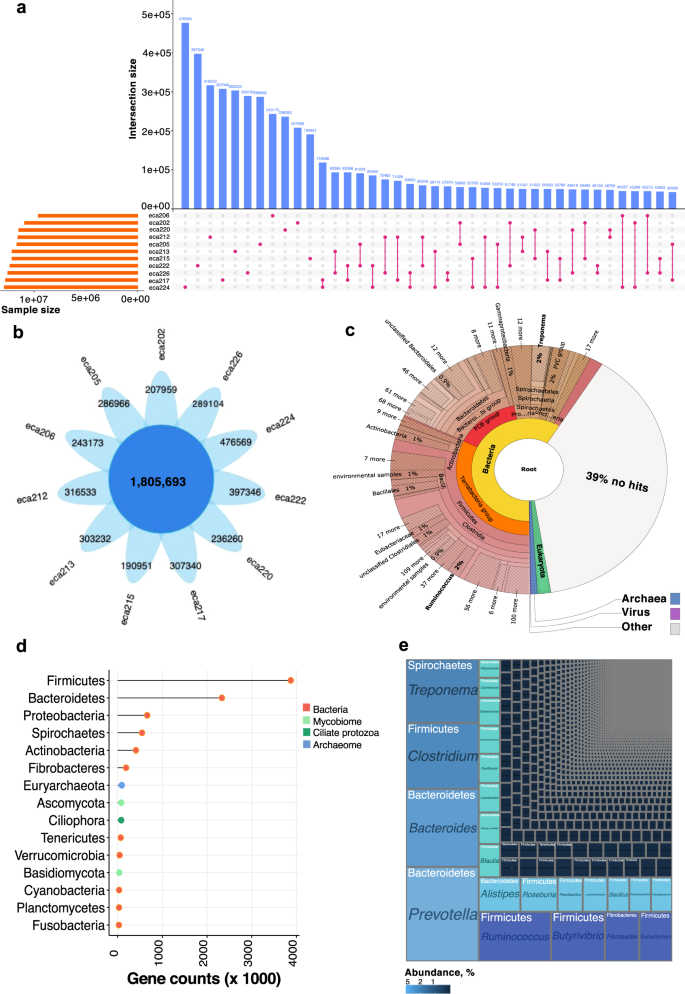
a Contribution of different sample sources (n = 11) to gene content of the horse gut microbial gene catalog. Vertical blue bars represent the number of genes present in only one sample or shared between pairs of samples. Horizontal orange bars in the lower panel indicate the total number of genes contained in each sample. b Flower plot showing the number of core genes shared between all samples (n = 11) and those specific for each individual. c Visualization of the taxonomic assignment of Illumina trimmed paired reads in a Krona plot using the software tool Kaiju. d Lollipop plot showing the gene counts identified by the Kaiju resolved at the phylum level. Dots are colored by kingdom. e Treemap showing the taxonomic ranking of the main taxa in the gene catalog using Kaiju. The size of each box is proportional to the number of genes assigned to each taxon. The placement of boxes is arbitrary concerning boxes within the same taxonomic rank and does not correspond to any form of phylogeny or relatedness.
Taxonomic annotation of the microbial gene catalog was implemented with Kaiju31 using a greedy mode and the NCBI nr_euk reference database. While 39% of the gene sequences were unclassified, this approach annotated 61% of the gene sequences (Fig. 1c) and revealed a diverse community of 95 phyla encompassing 1110 families and 4179 unique genera (Supplementary Note, Supplementary Data 3). More than 90% of these genera have not been previously described in horses. Among classified genes, bacteria (95.58% of genes, n = 2927 genera) defined most of the assemblage in terms of abundance and diversity, followed by a handful of eukaryotes (2.55% of genes, n = 1081 genera), and archaea (1.20% of genes, n = 170 genera). At the phylum level, genes related to bacterial phyla Firmicutes (47.1%) and Bacteroidetes (21.8%) greatly outnumbered Proteobacteria (6.0%), Spirochaetes (3.9%), and Actinobacteria (3.1%; Fig. 1d). Consistent with two recent metagenomic studies in horses30,32, Ascomycota (0.56%) and Basidiomycota (0.25%) were among the top annotated Eukaryota phyla in the gut. At the same time, Saccharomycetaceae represented the dominant eukaryote family30. Many genes in the catalog pertained to the Ciliophora phylum (n = ~80,000 genes), stressing the importance and central role protozoa likely play in host function and microbiome metabolism (Fig. 1d). The most observed protozoa were Stentor, Stylonychia, Paramecium, Ichthyophthirius, and Tetrahymena. The taxonomic analysis also revealed that the core group of genes was defined by Prevotella (5.10%) and Bacteroides (2.98%), both dominant in the gut of marathon and triathlon athletes, respectively33, along with Ruminococcus (3.17% of genes), Treponema (3.11%), Clostridium (2.70%), Butyrivibrio (2.42%), and Fibrobacter (1.37%; Fig. 1e; Supplementary Data 4). All of these fully agree with the core microbiota of horses inferred from 16S rRNA-based sequencing24,34,35,36.
To gain functional insights, we annotated genes via KEGG orthologous groups (KOs) and carbohydrate-active enzymes (CAZymes) using the non-supervised orthologous groups (EggNOG) database. Results revealed a total of 12,060 KOs and 137 unique CAZymes, which encompassed 44% (n = 11,132,404) and 3.38% (n = 665,235) of the gene catalog, respectively. Most KOs had essential microbial gut functions, including metabolism (transport and metabolism of amino acids, carbohydrates, nucleotides, and lipids), genetic information processing, and signaling (Supplementary Data 5). We also identified potential functions related to the biosynthesis of secondary metabolites, quorum sensing, and prokaryotic defense system, which help maintain the microbiome's structure and the host's health (Fig. 2a). Given the abundance and variety of carbohydrates in the horse diet, the functionalities of glycosidic bond processing enzymes were investigated using the CAZy database. The great majority of these CAZYmes pertained to glycosyl-transferase (GT, 26.8%, n = 228,878 genes) and glycoside hydrolase (GH, 64.9%, n = 554,661 genes) families, followed by carbohydrate-binding modules (CBM; ~4%, n = 37,474 genes) and polysaccharide lyases (PL) families (~1.6%, n = 13,801; Supplementary Data 6). Confirming what has been recently found in horses30, the most highly expressed CAZy family genes belonged to the GT2, GH5, and GH9 families, which contain diverse cellulases and hemicelluloses. Genes encoding GH13, GH57, GH77, and CBM48 families, characterized by their starch catalytic activities, were also highly represented in the catalog (Fig. 2b). Among 568,462 GHs and PLs genes, 53,569 (6.38%) were involved selectively in host glycan degradation (PL8, PL12, GH18, GH20, and GH3837; Supplementary Data 6).
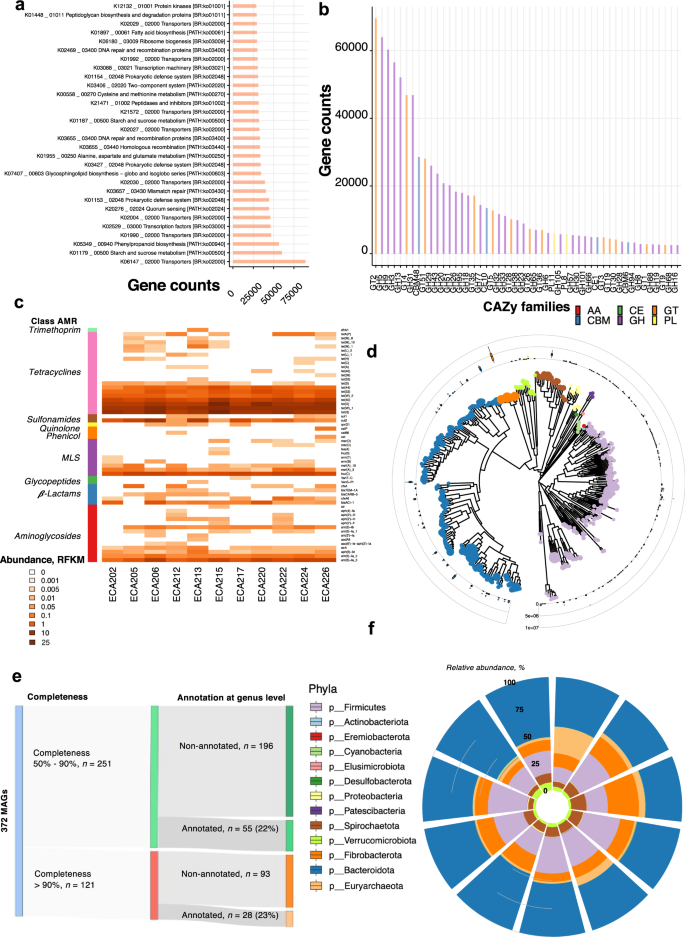
a Frequency of KEGGs pathways in the gut microbial gene catalog estimated from the orthologous groups (KOs). The horizontal bars represent the absolute number of genes found in each KEGG pathway. b The Frequency of total carbohydrate-active enzyme (CAZymes) families estimated in the gut microbial gene catalog. CAZymes are colored according to their class. c Heatmap shows each individual's (n = 11) normalized counts of antimicrobial resistance (AMR) genes based on the ResFinder database. The left column depicts the AMR class. d The Phylogenetic tree of the n = 372 MAGs detected. The outer cycle boxplots represent the abundance of each MAG. Boxplots are colored according to phyla. e Sankey diagram showing the numbers of MAGS with completeness between 50 and 90% or > 90%. Only a relatively small fraction of the MAGs (73 out of 372) was annotated at the genus level. f Circular stacked bar plot of the MAGs phyla abundance detected for each individual in the cohort (n = 11).
We subsequently investigated the presence of antimicrobial resistance (AMR) genes within our gene catalog. A total of 57 clusters of AMR genes representing the major antibiotic classes were observed, including tetracycline (n = 20), aminoglycosides (n = 14), and macrolides, lincosamides, and streptogramins (MLS, n = 9; Fig. 2c; Supplementary Data 7). The strong representation of Firmicutes and Bacteroidetes-associated tetracyclines resistance genes (tet(W), tet(Q), tet(O), tet(40)) and MLS resistance genes (lnu(C) and mef(A)) mirrored past observations in horses30 and matched that found in humans38, and livestock species such as cattle, pig, and chicken39,40,41. We detected an extended-spectrum of β-lactamase (ESBL) blaACI-1 found in several Negativicutes (Gram-negative Firmicutes) but rarely detected in animal or human gut microbiomes42.
Last, we built 372 non-redundant prokaryotic MAGs at >50% completeness and contamination ≤10% (Supplementary Note, Supplementary Data 8, Supplementary Fig. 2a–h). Among these, 121 MAGs were estimated to be near complete; MAGs in this subset had minimal contamination (≤ 5%) and high completeness (> 95%). According to the Genome Taxonomy Database Toolkit (GTDB-Tk)43, this MAG repertoire was assigned to 361 bacteria and 11 archaea, involving bacteria from the Bacteroidetes and Firmicutes phyla, followed by Spirochaetes, Euryarchaeota, Verrucomicrobia, Fibrobacteres, and Cyanobacteria phylum (Fig. 2d). However, only 83 and 41 MAGs were classified at the genus and species levels, respectively (Fig. 2e). The MAGs pertaining to the Cyanobacteria, Proteobacteria, and Verrucomicrobia phyla displayed variable abundances between hosts (Fig. 2f). This trend was bolstered at the lower taxonomic level, except for MAGs assigned to Fibrobacter spp. (Supplementary Fig. 2h). Most MAGs encoded enzymes that degrade polysaccharides (Supplementary Note, Supplementary Data 8, Supplementary Fig. 3).
Notably, most MAGs (n = 369) were new for horses44,45, increasing the mappability of metagenomes and expanding our understanding of the horse microbiomes.
The dominant basal gut phylotypes defined two taxonomical and functional microbial communities
We first profiled microbial taxa in each metagenomic baseline sample at the taxonomic (NCBI nr_euk reference database) and functional (EggNOG database) levels. Then, we retained the most dominant microbial phylotypes. These phylotypes harbored 1,146 unique genera (accounting for 95% of the classified sequences) with abundance and prevalence in the top 25% and 50% quantiles, respectively (Supplementary Data 9). They were represented mainly by prokaryotes (91%), a handful of eukaryotes (7.15%), and archaea (1.75%). In agreement with 16S rRNA gene sequencing data from the same samples, Prevotella, Fibrobacter, Clostridium Ruminococcus, and Treponema were the most prevalent genera in most individuals (Supplementary Data 10 and 11). However, all individuals harbored large amounts of uncharacterized taxa in the horse, such as Mediterraneibacter, Coprobacillus, Mucilaginibacter, Chitinophaga, Flavobacterium, and Enterocloster (Fig. 3a).
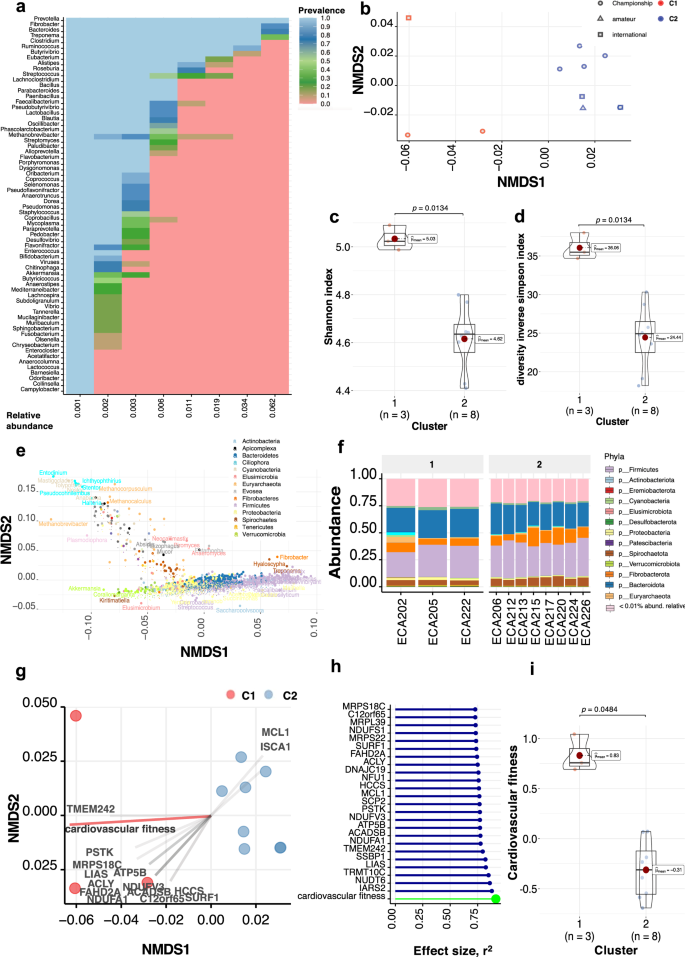
a Dominant phylome and its prevalence at different detection thresholds (relative abundance). The percentage of shared items and the proportion of shared samples are represented on the y- and x-axis. b NMDS ordination analysis (Bray–Curtis distance) of dominant phylotype composition. Points denote individual samples (n = 11), colored according to the clustering group. The shape of the dots indicates the competition level of horses. c, d Violin plot representing Shannon Diversity Index and inverse Simpson index of dominant phylotypes according to the clustering groups, respectively. In all cases, colors indicate community classification, cluster 1 (red color, n = 3) and cluster 2 (blue color, n = 8). Boxplots show the median, 25th, and 75th percentile, the whiskers indicate the minima and maxima, and the points lying outside the whiskers of boxplots represent the outliers. Adjusted p-values from two-sided Wilcoxon rank-sum test. e Biplot values of the discriminant phylotypes driving the NMDS ordination. The phylotypes contributing to the distinction between groups on at least one axis are depicted. Points are colored by phylum. f Taxonomic distribution of the dominant phylotypes grouped by phyla in each individual (n = 11). Individuals are split by cluster: cluster 1 (n = 3) and cluster 2 (n = 8). g NMDS ordination plot shows the covariates contributing significantly to the variation of dominant phylotypes determined by the envfit() function. The arrows for each variable indicate the direction of the effect and are scaled by the unconditioned r2 value. Dots represent samples (n = 11), which are colored according to the type of community: cluster 1 (red color, n = 3) and cluster 2 (blue color, n = 8). h Effect sizes of the main variables affecting the NMDS ordination. The length of the horizontal bars shows the amount of variance (r2) explained by each covariate in the model. Covariates are colored according to the type of dataset: athletic performance are in green and mitochondrial-related genes in blue. i Violin plot representing the cardiovascular fitness of each cluster, which was calculated as a composite of post-exercise heart rate, cardiac recovery time, and average speed during the race. Colors indicate community classification, cluster 1 (red color, n = 3) and cluster 2 (blue color, n = 8). The boxplots show the median, 25th, and 75th percentile, and the whiskers indicate the minima and maxima. The points lying outside the whiskers of boxplots represent the outliers. Adjusted p-values from the two-sided Wilcoxon rank-sum test.
The ordination analysis of these dominant microbial phylotypes yielded two distinct clusters of samples that recapitulated variation along the first axis (non-metric multidimensional scaling (NMDS), Bray–Curtis distances; Fig. 3b). The same pattern was supported by a permutational analysis of variance (pairwise PerMANOVA; p = 0.008, R2 = 0.3716). Cluster 1 individuals (n = 3 horses) exhibited higher α-diversity despite the small sample size (Shannon and inverse Simpson indices; p = 0.0134 for both, two-sided Wilcoxon rank-sum test, Fig. 3c, d). We then investigated which microbial taxa underpinned each cluster. Cluster 1 mainly included the enrichment of many poorly-characterized taxa (n = 318; IQR: 2.90e−04– 1.16e−04; Fig. 3e, f) that encompassed Proteobacteria (n = 132), Actinobacteria (n = 49), Verrucomicrobia (n = 17), Planctomycetes (n = 17), and Cyanobacteria (n = 7). Methanogens previously described in the bovine rumen46,47—including those belonging to the Methanomicrobiales (Methanothrix), Methanobacteriales (Methanosphaera, Methanobacterium, Methanothermobacter), and Thermococcales (Thermococcus) orders—were also more abundant in that cluster (DESeq2, adjusted p < 0.05; Supplementary Data 12). In contrast to the overwhelming diversity observed in cluster 1, a dwarfed biodiversity defined cluster 2 that assembled around highly efficient fiber degraders (members from the Lachnospiraceae taxa: Anaerostipes, Butyrivibrio, Blautia, Coprococcus, Dorea, Eubacterium, Hespellia Lachnospira, Oribacterium, Roseburia, and L-Ruminococcus) and downstream users of degradation products (Prevotella, and Treponema)47 (DESeq2, adjusted p < 0.05; Supplementary Data 12, Supplementary Fig. 4a–d).
The sample distribution based on microbial KOs (PerMANOVA; R2 = 0.3725, p = 0.008) and CAZymes (PerMANOVA; R2 = 0.6063, p = 0.005) echoed that of taxonomical composition, supporting taxa segregation by their underlying biochemical activities (Supplementary Fig. 5). Cluster 1 captured higher CAZyme diversity. It showed enrichment of CAZymes that might act in a concerted manner to cleave plant polysaccharides into fermentable monosaccharides (GH6, GH39, GH54, GH82, GH84, GH99) or host glycosaminoglycans (PL8, PL12, GH18, GH20, GH29, GH88, and GH95; DESeq2, adjusted p < 0.05; Supplementary Data 13, Supplementary Fig. 4d). Cluster 1 also had more CAZymes for host glycans degradation (14.15%) than members of cluster 2 (9.63%). In agreement, the latter presented an enrichment of KOs related to glycan biosynthesis and metabolism (K09953, K18770, K12309, K12551, K01137, K03276, K12985, K14459). Cluster 1 also exhibited large metabolic increases (n = 520 KOs) in amino acids metabolism (K12256, K15226, K03852, K08688, K18049, K21062), energy production, e.g., genes within lipid metabolism (K10781, K16795, K01795, K01049, K12309), propanoate metabolism (K11264, K01659 K00382 K01720), fructose and mannose (K19633) and pyruvate (K02594), as well as methane production (K00202, K00204, K00583, K05884; DESeq2, adjusted p < 0.05; log2FC|>1.5|, Supplementary Data 14). In accordance with protein catabolism, plasma iso-valerate—a precursor for muscle glycogen1—was also higher in cluster 1 individuals (p = 0.0121, two-sided Wilcoxon rank-sum test). These findings strongly suggest that cluster 1 horses had a larger substrate range. However, this highly functional complexity could not be captured at the MAG level (Supplementary Note, Supplementary Data 8, Supplementary Fig. 6a–d).
Cluster 1 metagenomes also had reduced representation of the host DNA in the raw sequencing data (p = 0.05, two-sided Wilcoxon rank-sum test, Supplementary Fig. 5, Supplementary Data 2), suggesting reduced epithelium disruption or intestinal inflammation, commonly observed in endurance athletes48.
Basal dominant gut phylotypes were closely associated with cardiovascular fitness in equine athletes
To tie together the basal dominant gut phylotypes with the horse performances, we investigated whether diversity, compositional and functional differences between clusters were driven by nutrient intake or any horse-centered parameter. Recorded dietary data showed that participants had a constant standard varied macronutrient diet on training and non-training days [mean ± SD (% dry matter (DM) basis): 53.7 ± 19.3 MJ/day, 939 ± 42 g/day of protein (11 ± 1.9%), 678 ± 20 g/day of fat (8 ± 1.6%), 2190 ± 706 g/day of hydrolyzable carbohydrates (26 ± 8.9%), and 2320 ± 877 g of fermentable fiber (27 ± 2.8%)]. Individuals were fed 8.43 ± 2.99 kg/day, with an 80:20 forage to concentrate ratio (Supplementary Data 1). None of the macronutrient intakes were statistically different between the two clusters (p > 0.05, two-sided Wilcoxon rank-sum test), and none of them were the major driving forces of the microbiome composition (PerMANOVA; R2 = 0.102 ± 0.089 and p-values ranging from 0.055 to 0.82; Supplementary Fig. 7a–d). Likewise, KOs (PerMANOVA; R2 = 0.034 ± 0.010 and p-values ranging from 0.940 to 0.969) and CAZymes profiles (PerMANOVA; R2 = 0.030 ± 0.016 and p-values ranging from 0.250 to 0.971) were also independent of nutrient intakes. Gut community composition was neither linked to horse sex (PerMANOVA; R2 = 0.2486, p = 0.266), or age of the athletes (PerMANOVA; R2 = 0.0952, p = 0.409; Supplementary Fig. 7e–g). Moreover, clusters 1 and 2 did not overlap with the horse kinship (Supplementary Fig. 8).
After confirming that these confounding factors did not define the gut microbiome phylotypes, we next tested if any host-centered omic or phenomics dataset, including transcriptomics, metabolomics, blood biochemical assay profiles, acylcarnitines, and cardiovascular fitness parameters, best captured the distributions of metagenomic samples (Supplementary Note, Supplementary Data 15–18, respectively). Cardiovascular fitness—a composite variable of post-exercise heart rate, cardiac recovery time, and average speed during the race—was the primary driver of the overall structural variation of the gut metagenome (envfit, R2 = 0.9192, adjusted p = 0.005; Supplementary Data 19).
This cardiovascular composite parameter aggregated 39.64% of fecal microbiome community variation, thereby outperforming the expression of several mitochondrial-related genes (Fig. 3g, h). Horses from cluster 1 had significantly higher cardiovascular fitness than cluster 2 members (p = 0.048, two-sided Wilcoxon rank-sum test, Fig. 3i). That did not increase blood lactate concentration (p = 0.921, two-sided Wilcoxon rank-sum test), which is a proxy for glycolytic stress and disturbance in cellular homeostasis1.
An independent validation set confirmed that Lachnospiraceae bacteria were negatively associated with cardiovascular fitness in highly trained equine athletes
We further attempted to validate this association with 16S rRNA sequence data from the gut microbiota of 22 independent highly trained endurance horses (Supplementary Data 20, 21)
As with the study cohort, the microorganisms' community profiles could be distinguished based on the horse's cardiovascular fitness (pairwise PerMANOVA on a Bray–Curtis distance matrix; L vs. H: adjusted p = 0.057, R2 = 0.109, Supplementary Fig. 9a, b), with higher microbiota dispersion in more fit athletes (betadisper(), L vs. H: adjusted p = 0.0243; Supplementary Fig. 9c). We used sparse Partial Least Squares-Discriminant Analysis (sPLS-DA) to find a taxa panel that discriminated between the more and less fit horses (Supplementary Fig. 9d). This analysis only supported the presence of Mogibacterium and the yet undefined members of Rhodospirillaceae, Enterobacteriaceae, Planococcaceae, and Sphingobacteriaceae families in more fit individuals but not the other 252 poorly-described bacteria members. This likely reflects an under-representation of these taxa in existing 16 S rRNA reference databases. Conversely, the sPLS-DA corroborated the enrichment of Clostridiales, Erysipelotrichaceae, and Lachnospiraceae taxa in less competitive individuals. Markedly, genera such as Blautia, Butyrivibrio, Coprococcus, Dorea, Desulfovibrio, Hespellia, Lachnospira, and L-Ruminococcus (all pertaining to the Lachnospiraceae family) were consistently depleted in less fit athletes in both the discovery and the validation sets. In addition, Dorea and the polyphyletic Ruminococcus were consistent hallmark genera of less competitive individuals (DESeq2, adjusted p = 0.0491; Supplementary Fig. 9e, f) across both discovery and validation cohorts. This further strengthens the association between the horse's cardiovascular fitness and gut microbiota composition.
Although larger metagenomic cohorts and improved reference collections are required to validate the relationship between athletic performance and gut microbiome, these data do, however, confirm the negative association between Firmicutes (notably Lachnospiraceae taxa) and cardiovascular fitness in endurance athletes.
Holo-omics: microbiomes with a higher prevalence of Lachnospiraceae taxa signed lower cardiovascular fitness and pointed toward likely impaired mitochondrial capacity
To further characterize the microbiome-host crosstalk and identify molecular differences between the two types of cardiovascular outcomes in elite horses, we integrated multi-omic datasets from the host and associated gut microorganisms through a multivariate matrix factorization approach (DIABLO; see "Methods"). To achieve this integrated perspective coined as holo-omics49, we combined host-centered omic and phenomics data with the fecal shotgun metagenomics, SCFAs composition, and the concentrations of bacteria, anaerobic fungi, and protozoa.
First, we observed strong covariation between the dominant phylotypes and the genetic functionalities derived from KOs (r2 = 0.99) and CAZymes (r2 = 0.98; Fig. 4a). This apparent correlation supports the added value of microbiome functions for status prediction rather than composition alone, as noted in human athletes21,50. Concomitantly, the microbiome composition highly covaried with the mitochondrial transcriptome (r2 > 0.80) and loads of fecal protozoa (r2 > 0.80; Fig. 4a). The targeted (acylcarnitine profiles and biochemical assays) and untargeted host metabolomic analysis (1H NMR) also accounted for the metagenome variation, albeit of lesser significance (r2 = 0.57–0.61; Fig. 4a). Then, to add biological meaning to the predicted model, we investigated the relationship between the DIABLO-selected features with the highest covariation (Supplementary Fig. 10a–c). The first latent variable of the predicted model indicated that athletes with higher cardiovascular fitness harbored a wide range of multi-kingdom and yet undescribed clades in horses (Fig. 4b). Consistent with the univariate analysis, it included the facultative bacterial predator Lysobacter, Akkermansia, generally regarded as health-promoting bacteria in athletes51,52,53,54,55, along with anaerobic fungi (Ophiocordyceps, Pseudogymnoascus, Trichoderma, Talaromyces, Rhodotorula, Exophiala, Puccinia), methanogens (Methanothermobacter, Methanothrix) and algae (Emiliania and Porphyra). It is worth noting that the ciliate protozoa, at up to 18% of the biomass (~109 cells/g of stool), were discriminative of more fit individuals (Supplementary Fig. 10d). Conversely, less fit subjects were largely defined by members of the family Lachnospiraceae, Treponema, Prevotella, and other commonly described taxa in horses, including Clostridiales and undescribed Erysipelotrichaceae taxa. Paired with this less complex community, the first latent variable pointed at a redundant functional diversity, spanning CAZymes to extract energy from recalcitrant polysaccharides (GH8, GT36, GH51, GH28, GT2, GH5, GH3; Fig. 4c). While intestinal microbiota members belonging to the Lachnospiraceae family are known to produce acetate and butyrate56, none of these SCFA were significantly increased in the feces or plasma of these athletes (p > 0.05; two-sided Wilcoxon rank-sum test), and the fecal pH remained unchanged (p = 0.837; two-sided Wilcoxon rank-sum test; Supplementary Data 22).
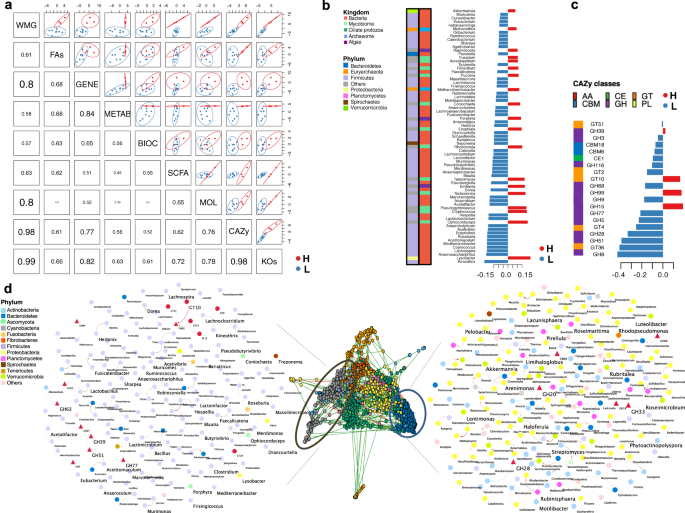
a Matrix scatterplot showing the correlation between the first components related to each dataset in DIABLO according to the input design. b Dominant microbial genera contributing to the separation along with component 1 of the microbiome dataset. Microbiome data are centered log-ratio-transformed, and bar length indicates loading coefficient weight of selected phylotypes, ranked by importance, bottom to top. Columns on the left depict the kingdom and phylum of each discriminant phylotype. c CAZymes contributing to separation along with component 1 of the CAZy dataset. CAZymes profiles are log-transformed median-scaled values. Bar length indicates the loading coefficient weight of selected CAZymes, ranked by importance, bottom to top. In all cases, colors indicate community classification, cluster 1 (red color, n = 3) and cluster 2 (blue color, n = 8). The column on the left depicts the CAZymes class. d Co-occurrence network analysis of dominant phylotypes and carbohydrate-active enzymes (CAZy) types datasets using sparse inverse covariance estimation for ecological association inference (SPIEC-EASI). Louvain clustering was able to generate 12 feature co-occurrence modules. The two extreme assortative modules are depicted in detail using Cytoscape. A positive correlation between nodes is indicated by red connecting lines, and a negative correlation by blue. A circle or triangle denotes microbial clades and CAZymes features. Microbial nodes are colored by phyla. Elements with larger text sizes are those revealed as discriminant along with component 1 by the DIABLO approach. Edge width corresponds to the strength of the association between features.
At the transcriptome level, the first latent variable supported an impairment rather than improved metabolic flexibility in the less fit individuals. The latter exhibited downregulation of mitochondrial-related genes involved in β-oxidation (ECI1, SCP2, ACLY), electron transport chain (TMEM242, NDFB4, TMEM126B, NDUFV3, NDUFA1, NDUFA10, SURF1, NDUFV1, DLD), Ca2+ translocation (PMPCA, VDAC2, PHB), mitophagy (TOMM40), and biogenesis (SSBP1, ACSS2; Supplementary Fig. 10e). These results suggested decreased mitochondrial fatty acid oxidation and increased glucose catabolism, which progressively impedes longer running times, and fatigue resistance. In agreement with this notion, less fit individuals presented reduced concentrations of glucose (p = 0.0484, two-sided Wilcoxon rank-sum test), increased accumulation of long-chain acylcarnitines in plasma (e.g., oleoyl carnitine, p = 0.0484; hydroxy oleoyl carnitine, p = 0.0242, two-sided Wilcoxon rank-sum test) and a tendency for augmented non-esterified fatty acids (NEFAs) in plasma (p = 0.0848, two-sided Wilcoxon rank-sum test). Therefore, less diverse microbial communities dominated by a few Firmicutes-derived families (mainly Lachnospiraceae) could constrain the horse mitochondrial aerobic ATP production for extended cardiovascular fitness.
Frenemies: Lachnospiraceae and poorly-described phylotypes in horse athletes
To gain insight into the co-occurrence and co-exclusion relationships between multi-kingdom microbial genera and functions, we applied an inverse covariance estimation for ecological associations. This approach identified 12 modules. Among them, we uncovered two extreme assortative modules characterized by robust microorganism-microorganism or microorganism-functions interactions (Fig. 4d) that agreed with the other univariate and multivariate analytical frameworks. The first module was mainly characterized by bacterial interactions within the Firmicutes phylum (mainly from the Lachnospiraceae family) and CAZy families that can target the substrate of plant structural polysaccharides (GH3, GH39, GH51, GH82, GH84). On the other hand, the second extreme network encompassed widespread yet undescribed bacteria in horses from the Proteobacteria, Actinobacteria, Planctomycetes, and Verrucomicrobia, together with CAZymes active in the degradation of the complex structure of plant cell-wall materials (GH28) and host glycans (GH20, GH18, GH33; Fig. 4d).
Discussion
The current study presents a comprehensive horse gut microbiome gene catalog and its association with endurance performance. The more than 25 million non-redundant genes in this catalog have widened 20-fold the number of genera known to reside in the gastrointestinal tract of horses20,32,44,45,57,58, uncovering a substantial number of bacteria (n = 2927), archaea (n = 174), and Eukaryota (n = 1081). This catalog also captured a wide array of functions related to complex carbohydrate fermentation and functional capacities that enabled more energy extraction from dietary, microbial, and host resources, which likely provided a performance advantage for athletes. For example, genes encoding enzymes for glycosidic-bond cleavage (GHs and PLs) represented the majority (67%) of the CAZyme genes, highlighting the indispensable role of gut microorganisms in glycan metabolism. Intriguingly, 6.38% of GHs and PLs were involved selectively in cleaving endogenous host-related glycosylated proteins and glycans37. It is yet unclear whether these CAZymes correspond simply to the peptidoglycan-processing machinery required for bacterial cell growth and division or whether peptidoglycan and highly glycosylated proteins represent a major carbon source for some bacteria or the host after being transformed to acetate59.
In addition, we have identified a set of 372 MAGs. The number of microbial genes and MAGs will likely increase as more samples are analyzed, as seen in equids44,45 and other livestock gut metagenomes29,60,61,62. Nonetheless, the present metagenomic genes and MAGs repertoire are a step forward in explaining the composition and function of the horse gut microbiome, especially in the context of endurance exercise.
Along with fatigue resistance, cardiovascular fitness is a crucial indicator of endurance performance in human athletes9. In the considered elite endurance horses, the cardiovascular capacity defined about ~40% of the variation in the most dominant gut phylotypes (e.g., most abundant and ubiquitous) in the absence of any confounders. This association was replicated in an independent cohort of elite horses and outweighed that estimated between marathon runners' cardiovascular fitness and their microbiota (~22% of explained variance22). Moreover, we found that higher cardiovascular capacity was mostly associated with uncharacterized microbial features and functional pathways. Cardiorespiratory fitness matched increased gut microbiota diversity in marathon runners21. A possible explanation for this phenomenon could stem from the broader use of resource compounds, including SCFAs, host glycans, dietary and microbial proteins, complex lipids, propanoate, pyruvate, and hydrogen, to produce higher relevant goods for host energy requirements. This notion was exemplified by the extensive enrichment of CAZymes and KOs involved in the metabolism of glycans, amino acids, carbohydrates, and lipids.
The deep phenomics applied to these horses pointed to the microbiome-mitochondria crosstalk as a potentially highly effective way to modulate cardiovascular capacity. Conceptually, the higher availability of goods likely influenced the mitochondria function and readouts. This hypothesis paralleled increased glucose availability in serum, a precursor for glycogen in muscle, alongside reduced circulating acylcarnitines and NEFA levels. The acylcarnitine and NEFA drop was possibly not due to reduced substrate availability but to an increase in transport into the mitochondria through the carnitine shuttle, typically for ß-oxidation. In concert, mitochondrial-related genes involved in energy resilience, biogenesis, and Ca2+ cytosolic transport were simultaneously upregulated in participants with greater cardiovascular capacity suggesting enhanced mitochondrial oxidative phosphorylation and FAO, the two metabolic pathways central to energy pr...
Comments
Post a Comment