Spatial transferability of an agent-based model to simulate Taenia ... - Parasites & Vectors
Overview of approach
As a first step we develop and introduce a new module into the ABM capable of representing the process of pig development of antibodies upon exposure to T. solium eggs or through maternal transmission. We then used existing datasets from two distinct previously published community intervention trials to calibrate the model and then to assess its transferability. The revised model was calibrated, following the non-local approach described above, using observed data of human taeniasis prevalence (HTP), pig cysticercosis prevalence (PCP) and pig seroincidence (PSI) obtained from eight rural villages located in the Piura region in northern Peru. The resulting calibrated parameters was then transferred to a second group of 21 geographically separate destination villages located in the same department. Baseline values and intervention impacts were simulated, and the pig seroprevalence data collected during an intervention trial in the destination villages were used to quantitatively validate both the spatial transferability of the model and the effect of control interventions simulated by the model.
Model description
ABM short description
CystiAgent is an ABM that simulates T. solium transmission in a rural village. The model will be briefly described below while the details of the core structure are described elsewhere [18]. The model incorporates humans, pigs and households as agents, capturing the essential factors for T. solium transmission. It considers the geographical distribution of households, human and pig populations, and their behaviors. Human agents are associated with outdoor defecation sites around their households, which become contaminated if they carry a tapeworm. The degree of contamination depends on the availability and use of latrines. Pigs can become infected by coming into contact with contaminated sites within their roaming areas. The model calculates the number of T. solium cysts in infected pigs based on the level of contamination they are exposed to.
Both the human and pig populations are dynamic, reflecting natural rates of births, deaths, emigration and immigration. New human agents are periodically introduced to simulate migration or visits. The pig population aligns with observed herd size, slaughter age and import/export patterns in the region. Each household manages its herd size through export, sale or slaughter. When a pig is slaughtered at home, the resulting pork portions are distributed among the members of the household owning the pig as well as neighboring households, and subsequently consumed. Ingesting a T. solium cyst is associated with a probability of developing a tapeworm, and multiple tapeworm infections are not allowed. The model does not consider human cysticercosis or related seizure disorders. Minor modifications to the CystiAgent core are described in Additional file 1.
Pig seroconversion module
To compare the simulated and observed values of PSI, a new module was added to the previous version of CystiAgent [18] to simulate the pig seroconversions process. In the model, a pig is considered seropositive if it has developed a level of antibodies that would result in a positive EITB assay according to the experimental setup described above. As depicted in the flow diagram in Fig. 1, the model accounts for four potential causes of serological state change in pigs: infection, exposure to T. solium proglottids or eggs and transfer of maternal antibodies.
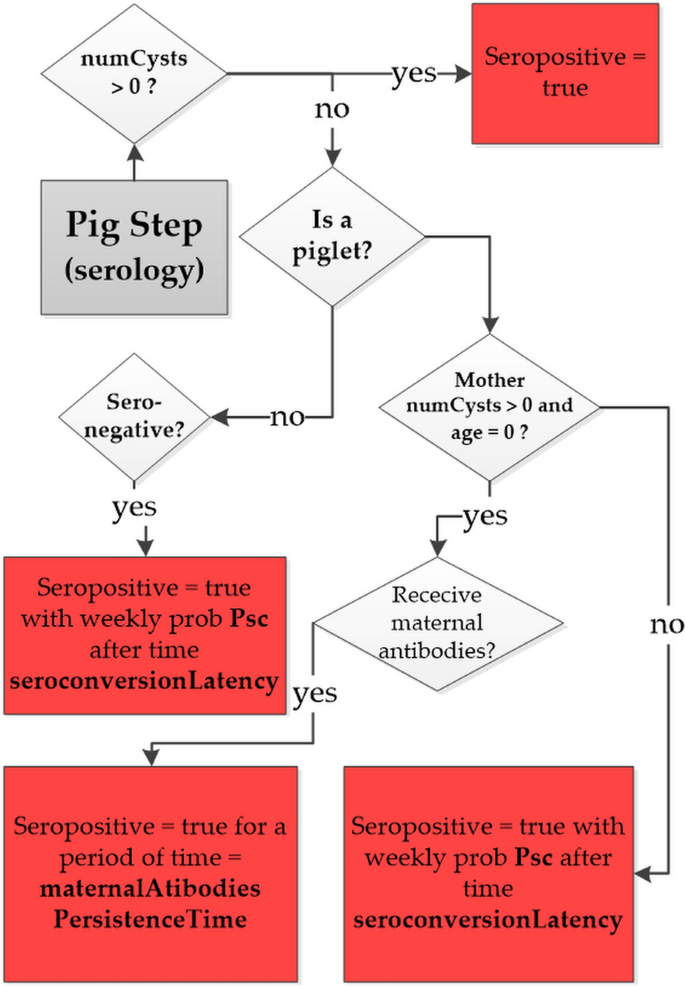
Flow diagram representing the process of development of antibodies against T. solium in pig. In the diagram numCysts is the number of cysts infecting the pig and Psc is the probability of seroconversion
The seroconversion is reversible only in the latter case, and the piglet returns to a seronegative state following weaning unless it has become positive through infection or exposure. The process of seroconversion through exposure is based on the calculation of T. solium contamination levels to which the pig is exposed during each model time-step (1 week). As described in [18], the levels of contamination from proglottids CP and eggs CE are assumed to be proportional to the number of defecation sites contaminated with proglottids and eggs within the pig roaming area, respectively. The factor seroConvertPtoEFact accounts for the lower density of T. solium eggs in a site contaminated with eggs compared to a site contaminated with proglottids. For each simulation time step and for each pig agent, the model counts the number of defecation sites contaminated with proglottids, eggs or both to which the pig is exposed. The resulting contamination levels are then inputted into an exponential dose-response model to calculate the weekly probability of seroconversion (Psc(t)) for each pig during the timestep t:
$$\mathrm{Psc}\left(t\right)= 1- {e}^{-\mathrm{seroConvert} \bullet \mathrm{CT}\left(t\right)},$$
(1)
where \(\mathrm{CT}\left(t\right)=\mathrm{ CP}\left(t\right)+\mathrm{seroConvertPtoEFact }\bullet \mathrm{ CE}(t)\) represents the total contamination to which the pig is exposed during the time step t and seroConvert is the exponential dose–response model factor. If exposure results in seroconversion, the pig's serological state changes to seropositive after a latency period of 2 week (as specified by the model parameter: "seroconversionLatency"). The process of seroconversion through maternal antibody transfer is represented as follows: when piglets are born to a PC-infected sow, they acquire maternal antibodies and become immediately seropositive with a probability specified by the model parameter "propPigletsMaternalProtection," regardless of the mother's cysts burden. The piglet's seropositive status is maintained for a period of 14 weeks [23], after which the piglet seroconverts to seronegative. During this period, the piglet is exposed to T. solium environmental contamination and, like any other pig agent, can seroconvert through exposure with a weekly probability \(\mathrm{Psc}(t)\). The parameters introduced in the seroconversion module of CystiAgent are presented in Table 1. Three of these parameters were selected for the purpose of calibrating the model.
Origin and destination field trial datasets
In this study, two separate datasets are used. The first dataset (referred to as the "origin dataset") serves as the calibration set, while the second dataset (the "destination dataset") is used to evaluate the spatial transferability and the simulated intervention effects. These datasets stem from two distinct community cluster randomized trials conducted in rural villages situated in two separate areas of the region of Piura in the northwest region of Peru (Fig. 2). In these rural communities, pigs are allowed to roam freely, access to proper sewage and latrine systems is limited, and open defecation practices are prevalent, leading to exposure of pigs to human feces and hence to the creation of an optimal environment for the transmission of T. solium [11].
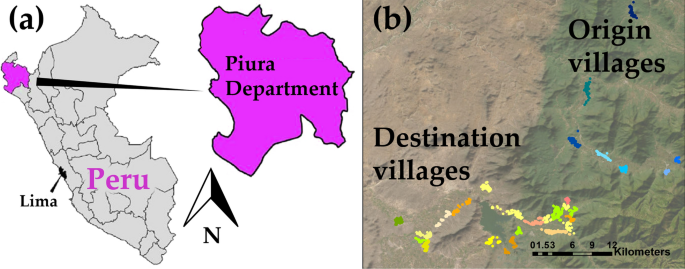
Study area maps; a the Piura region in north Peru; b the relative location of the 8 origin villages and the 21 destination villages. Each dot on the map represents a different household. Households from the same village are in the same color. Households from origin villages are showed in shades of blue while the households from destination villages are represented in shades of green, orange and yellow
The origin trial [4, 5], conducted between 2014 and 2015, aimed at evaluating the impact of education and increased community surveillance on T. solium transmission in eight villages. The trial resulted in a comprehensive household census and in the collection of blood samples from all pigs older than 6 weeks over four rounds. At the end of the trial, the HTP and PCP were determined through mass human stool screening and pig necroscopy, respectively. No significant effects of increased community surveillance were observed at the end of the trial period, so all the data collected can be considered representative of the baseline endemic level of all enrolled villages.
The destination dataset is derived from a field control intervention trial conducted from 2015 to 2017 [8]. Out of the 23 villages included in the trial, data from 21 villages were used to create the destination dataset, while the remaining two villages were excluded because of the small pig population (7 pigs) or the absence of significant seroprevalence at the baseline pre-intervention stage (no transmission ongoing) [25]. Analogously to the origin trial, a baseline household census was conducted to gather information relevant to T. solium transmission. The villages were randomly assigned to six different intervention arms each with a different T. solium control strategy. Three of these arms focused solely on human intervention: Mass Treatment (Mass Trt), Ring Treatment (Ring Trt) and Ring Screening (Ring Scr). The remaining three arms targeted both humans and pigs: Mass Trt (P), Ring Trt (P) and Ring Scr (P). In the Mass Trt strategy, all village residents ≥ 2 years old received a single dose of niclosamide every 6 months for a total of five treatment rounds. The Ring Trt involved conducting an active surveillance based on pig tongue inspection [26, 27], every 4 months for seven rounds. If cysticercosis was found in the tongue of a pig, a 100-m treatment ring was established around the household owning the pig. All members ≥ 2 years old from households within the ring were treated with two oral doses of niclosamide separated by 15 days. All the tongue-positive pigs were either purchased or treated with a single dose of oxfendazole. In the Ring Scr approach, the same active surveillance based on pig tongue inspection was conducted. Upon identifying a ring, stool samples were requested from villagers ≥ 2 years old residing within the ring and tested for Taenia spp. eggs or antigens. Individuals diagnosed with HT were offered a single dose of niclosamide treatment. The Mass Trt (P) strategy added pig treatment to human treatment with seven rounds of mass pig treatment occurring every 4 months. In Ring Trt (P) and Ring Scr (P) strategies, only pigs aged > 5 weeks within the identified rings were treated. The primary outcome of the trial was the determination of the PSI in all pigs aged > 5 weeks and born during the two year study through 7 pig serosurveys conducted every 4 months [8]. The secondary outcome of the study was HTP, which was determined offering presumptive niclosamide treatment at the study end to all residents ≥ 2 years old and collecting and testing stool samples from all treated individuals.
Pig serological outcome data were based on the enzyme-linked immunoelectrotransfer blot (EITB), which detect antibodies against T. solium cysticercosis in pigs [28]. Reaction to any of six glycoproteins, GP39/42, GP24, GP21, GP18, GP14 or GP13, was considered a positive result. The GP50 band was not considered because of the known cross-reaction with Taenia hydatigena, a co-endemic and highly prevalent infection of pigs [29].
Model calibration
The CystiAgent model was calibrated and validated based on empirical observations of HTP, PCP and PSI as target data in this study. In the model there is not an explicit representation of immunity in pigs. As result, pig seroconversion has no impact on the transmission process. Hence, as illustrated in Fig. 3, the calibration process was divided into two distinct stages: transmission calibration followed by seroconversion calibration. Model parameters were separated in two categories: local and non-local. As shown in Table 2, six non-local parameters were selected as calibration parameters. These parameters are connected with the processes of human and pig infection and pig seroconversion. Table 2 also shows the local parameters that vary from village to village in this study. The rest of parameters were kept constant across different villages.
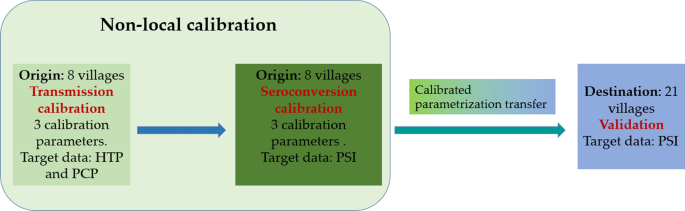
Model calibration and validation scheme
We applied the sequential Monte Carlo (SMC) approach to the likelihood-free method of approximate Bayesian computation (ABC), as described in [18, 30] for model calibration. We used a non-local approach to calibration [18]. The SMC method involves generating a sequence {εi} of decreasing ABC tolerances. For each tolerance εi, a straightforward ABC rejection sampler is executed, followed by exploration of the parameter space using importance sampling guided by the posterior distribution obtained in the previous stage. The initial rejection sampler begins with a uniform prior distribution of calibration parameters, spanning a wide and reasonable range of parameter values (see Additional file 1). The ABC distance function was defined as a normalized Euclidean distance between the observed and simulated vectors of target values for the 8 calibration villages [18]. During the calibration process, the first ABC-SMC stage involved 120,000 sampling points each corresponding to a different combination of calibration parameters. The second stage used 60,000 sampling points, the third stage used 40,000 sampling points, and the fourth stage used 30,000 sampling points. From the first to the fourth stage, only one simulation per sampling point was performed. The tolerance of each stage was set to accept the 20 sampling points that produced the lower values of the distance between observed and simulated vectors of target data. This tolerance was chosen as a trade-off between the convergence speed of ABC-SMC and the accuracy of sampling in the calibration vector space. In the fourth stage, both for transmission and seroconversion calibrations, the process converged as the distance between observed and simulated data for the best performing sampling point did not decrease compared to the third stage. To mitigate incorrect acceptance of sampling points due to the fluctuations of model output, after the first four stages we conducted six additional ABC-SMC rounds of 5000 points each. In these rounds, for each sampling point the simulations were repeated eight times and the average of the results from these eight repetitions was used to calculate simulated target data.
Estimation of empirical errors in the destination dataset and assessment of model transferability
The model was considered to have been successfully transferred to a destination village if the simulated value of PSI for that village fell within the error interval defined by twice the standard deviation (SD) of the time series of observed PSI values in that village. A direct calculation of PSI variance for the destination dataset is not feasible because of the absence of longitudinal observations in this trial. However, it is possible to hypothesize that intra-village variances are homogeneous across both the origin and destination villages, provided that this homogeneity holds true for the origin villages. This hypothesis of intra-village variance homogeneity across the origin villages can be tested using a Fligner-Killen test [31]. The resulting p-value of 0.722 indicates that intra-village variances are indeed homogeneous across the origin villages. To estimate the intra-village variance from the entire origin dataset, we considered the following random effect model:
$${\mathrm{PSI}}_{\mathrm{Vp}}= \mu +{U}_{\mathrm{V}}+{W}_{\mathrm{Vp}}.$$
(2)
The observed \({\mathrm{PSI}}_{\mathrm{Vp}}\) of village V is expressed as the sum of µ that is the average PSI value across all villages, UV a random effect to account for inter-village variability and WVp a random term accounting for intra-village variability due to individual pigs. The corresponding total variance σVW will be expressed as:
$${\sigma }_{\mathrm{VW}}= {\sigma }_{\mathrm{V}}+{\sigma }_{\mathrm{W}},$$
(3)
The variances σV of the UV term and σW of the WVp term represent the inter- and the intra-village variances associated to empirical PSI, respectively. This formulation assumes that the measurements of porcine cysticercosis at successive time points are uncorrelated, which may not be the case if the time interval between measurements is too short. If the time points are correlated, then we are underestimating the standard deviation. The random model was fit using the lmer module of the R package lme4 [32].
Simulation of spatially targeted interventions
As noted, CystiAgent explicitly represents the geographic space of simulated communities. As a result, not only can the observed spatial clustering of the parasite [16] be replicated by the simulations, but the model also allows for the simulation of spatially targeted interventions aimed at reducing the parasite burden in selected transmission hotspots. This is essential for simulating the effect of interventions based on the ring strategy adopted in the destination dataset. The ring interventions are simulated by the model exactly replicating the experimental protocol described in [8]. For Mass Trt and Mass Trt (P) interventions, humans and pig agents in the appropriate age segments were selected at random in the village and then treated. For Ring Trt, Ring Trt (P), Ring Scr and Ring Scr (P) intervention strategies, pigs were selected at random for tongue inspection (see the Table 3 for tongue inspection sensitivity and specificity used in the model). For each tongue-positive pig, a 100-m ring was opened, and humans were screened and/or treated accordingly. The efficacies of human niclosamide and pig oxfendazole treatments used in the model are shown in Table 3. The participation rates for pig tongue inspection, human stool screening and human treatment were set to be equal to the corresponding participation rates in the field trials for each round of each village.
Model software and simulations setup
The model was implemented using the MASON platform [36], a free, Java-based, discrete-event multi-agent toolkit, together with our in-house Java code for ABC calibration. Each village simulation started with 3500 burn-in time steps. The results reported in this study were obtained from 128 repeated simulations for each parametrization of the model. Within each simulation, the observables of interest were sampled 100 times at discrete points in time with a separation of 100 weekly time step. The simulations were run on the Exacloud Cluster at the Oregon Health and Science University Advance Computing Center, USA.
Comments
Post a Comment